Audrys G Pauza
Department of Physiology, FMHS
'in caHOOts’
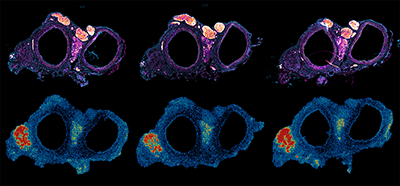
Spatial transcriptomics data from 10um-thick transverse sections of rat external and internal carotid arteries - description below.
The results of the 2025 image competition were announced at the BIRU Mini-Symposium on Tuesday 25 November 2025. The winner of the Hilary Holloway Prize and other category winners can be viewed below.
Click on each image to see a larger version.
See the highly commended entries here.
'in caHOOts’
Spatial transcriptomics data from 10um-thick transverse sections of rat external and internal carotid arteries - description below.
Spatial transcriptomics data collected using BGI STOmics (Stereo-seq) kit
10um-thick transverse sections of rat external and internal carotid arteries close to the common carotid artery bifurcation
TOP row: Sections labelled by immunohistochemistry against tyrosine hydroxylase (TH; magenta heat gradient) and DAPI (light blue). Labelled by TH, the carotid body (primary chemosensory organ monitoring arterial blood for oxygen saturation) is visible in between the lumens of the two arteries. Bright blobs at the top are sympathetic nerves transversing the bifurcation.
BOTTOM row: a heatmap pseudoimage depicting the number of mRNA transcripts (MID) detected in the tissue section shown above. Carotid bodies (in between the arteries) and sensory neurons in vagal sensory ganglion (in bottom left) stand out due to greater level of transcription and mRNA content compared to surrounding tissues. The bottom row is a synthetic image, i.e. a render of the number of transcripts (MID) detected on the spatial transcriptomics chip in specific locations. As coordinates on the chip are encoded by unique identifier sequence - this allows to match a specific area on the chip (used to acquire an image captured under the microscope) to reads generated by next-generation sequencing, encompassing spatial transcriptomics workflow.
Instrument and software used: Original images were acquired on Olympus VS200 using 10x air objectives at the BIRU. Widefield fluorescence image was captured as 16-bit and pseudocolours applied and adjusted in FiJi/ImageJ. Widefield images were deconvolved for 25 itterations using Lucy-Richardson algorithm implemented in DeconLab2 plugin in Fiji/ImageJ. Bottom plots were produced by lysing the tissues show at the top to capture mRNA contained in the tissue. Captured mRNA was used to generate short-read RNA sequencing libraries that were sequenced on MGI G400 instrument at Grafton Clinical Genomics. Following bioinformatics of the reads generated, synced image and read data were visualised using StereoMap software.
For me, this image reminds me of a bunch of owls, artery lumens appearing as eyes and the carotid body corresponding to the beak, that are in ca-HOO-ts (because that’s what an owl says!).
‘Portrait of a Monster: Mapping Osteoarthritis at the Molecular Level’
This matrix-assisted laser desorption/ionisation (MALDI) imaging mass spectrometry (IMS) ion image was collected as part of a spatial lipidomics study to characterise lipids in human osteoarthritic cartilage.
The image was acquired using the Bruker MALDI timsTOF flex mass spectrometer at a spatial resolution of 20 μm from a 10 μm tissue section of native undecalcified cartilage-on-bone.
MALDI-IMS visualisation was performed using SCiLS Lab 2026 Pro. Two ions are featured: red represents m/z 337.1915 and green represents m/z 739.3471, annotated as phosphatidylinositol 28:7.
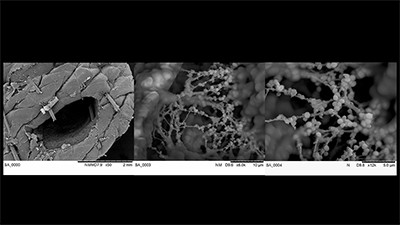
‘Exploring an Infected Wound’
Porcine skin infected with Staphylococcus aureus, an opportunistic bacterial pathogen. Sequential images show the wound region at low magnifications to clusters of spherical S. aureus within the wound at higher magnifications.
Instrument: Hitachi TM3030Plus tabletop SEM
Image processing: FIJI/ImageJ
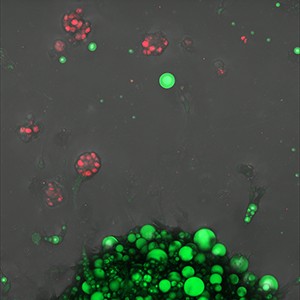
‘Kindling: adisphere ignites the tumour edge’
At the lip of an adisphere, cancer flares red. Cells caught in the act of rapid growth where closeness becomes catalyst. The scene reads like a spark meeting tinder... metabolism and malignancy in vivid, proximate dialogue.
Microscope: Zeiss LSM 800 Confocal, 10x objective
Software: Fiji/ImageJ; maximum-intensity projection
Sample preparation - Janneke Grundemann, Dr Emma Nolan